Automated 3D Voxel-Grid Reconstruction of Plants from Multi-view Image Sequences Contribute in the Largescale 3D Phenotypic Study Regulated by Genotypes
Collaborator(s): Sruti Das Choudhury, Srikanth Maturu , Ashok Samal , Tala Awada
Welcome to Pocket Science: a glimpse at recent research from Husker scientists and engineers. For those who want to quickly learn the "What, " "So what" and "Now what" of Husker research.
What
The complex interaction between a genotype and its environment determines the observable phenotypic characteristics of a plant that influence resource acquisition and yield. Image-based plant phenotyping analysis facilitates the computation of phenotypes without damaging the plants. It is an emerging research field that aims to link intricate plant phenotypes to genetic expression for global food security under dwindling natural resources and climate variability. The architectural complexity of plants increases over time due to variations in self-occlusions and phyllotaxy, i.e., arrangements of leaves around the stem. One of the central challenges to computing phenotypes from 2D images of plants (which is 3D in nature), especially at the advanced vegetative stage in presence of self-occluding leaves, is that the information captured in 2D images is incomplete, and hence, the computed phenotypes are inaccurate.
So What?
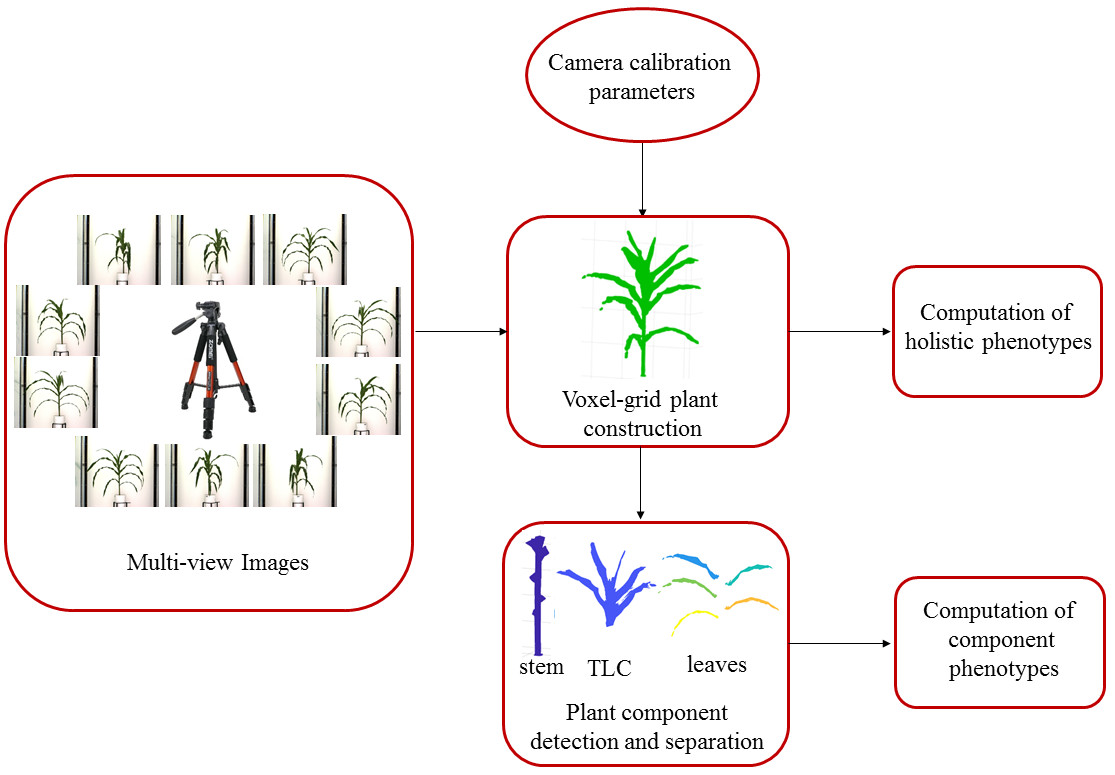
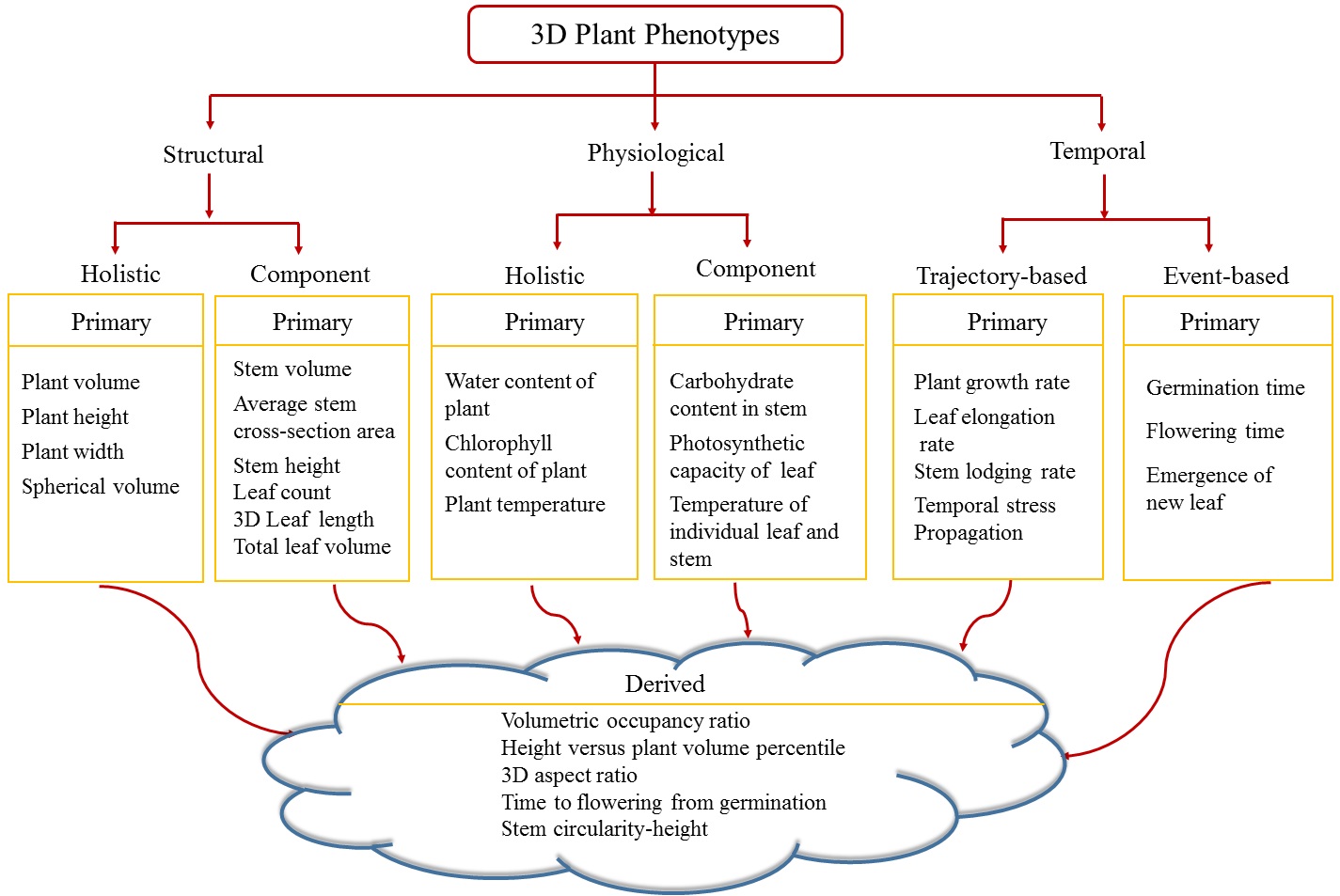
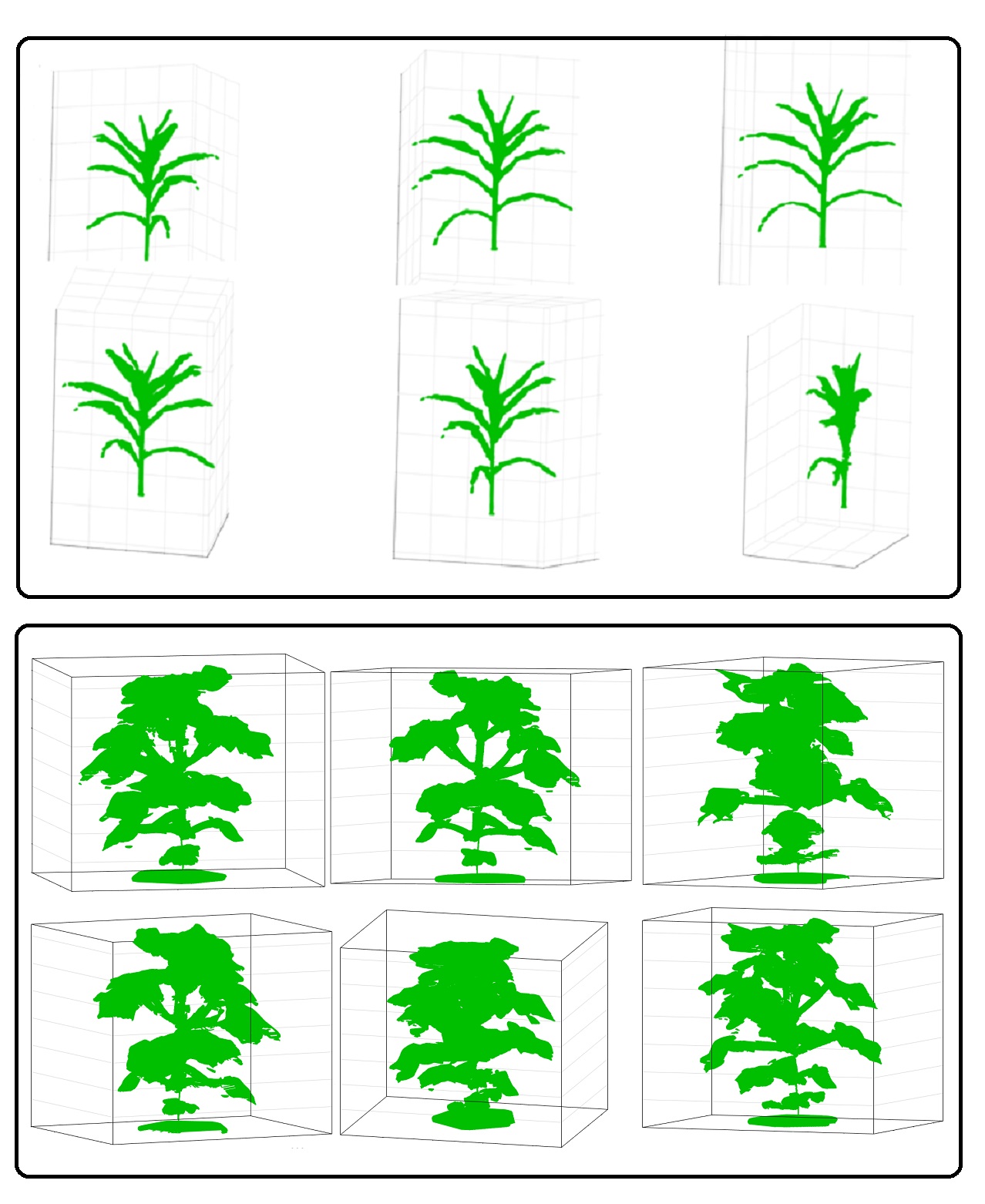
Dr. Sruti Das Choudhury, led the development of a novel method called 3DPhenoMV for computing 3D plant phenotypes (holistic and component) based on a voxel-grid reconstruction approach using multi-view visible light image sequences captured in an automated high throughput plant phenotyping platform where the distance between the pot and the camera is significantly larger (5.5 meters) compared to the state-of-the methods. 3DPhenoMV aims to achieve the fully automatic 3D model reconstruction of a large number of plants of various architectures at their late vegetative stages without requiring any manual intervention on an individual plant basis. The team also introduces a comprehensive taxonomy of 3D phenotypes, first of its kind, which addresses some of the pitfalls associated with 2D images due to self-occlusions and leaf crossovers. To evaluate the proposed method, a publicly available benchmark dataset called the University of Nebraska-Lincoln 3D Plant Phenotyping Dataset is released to foster research in 3D image-based plant phenotyping, facilitate new algorithm development, and enable uniform comparisons among competing methods.
Now What
This scalability and lack of human interaction will contribute to the method’s adaptability for a large scale phenotypic study regulated by genotypes, and also the method can be applied in the study of quantitative genetic engineering to identify loci controlling variation in the 3D phenotypic traits. Future work will consider creation of a new dataset consisting of larger number plants belonging to different genotypes with additional views, and validation of the phenotypes with physical measurements from the plant, destructively if needed, e.g., for biomass. The method will also be evaluated to estimate the temporal variation of phenotypes controlled by genetic factors for heritability analysis of many other species.
Publication: https://www.frontiersin.org/articles/10.3389/fpls.2020.521431/abstract